Leveraging Sparse Annotations for Leukemia Diagnosis
on the Large Leukemia Dataset
**Department of Hematology, Chughtai Lab , Lahore, Pakistan,
***Center for Research in Computer Vision, University of Central Florida, United States
Abstract
Leukemia is 10th most frequently diagnosed cancer and one of the leading causes of cancer-related deaths worldwide. Realistic analysis of Leukemia requires White Blook Cells (WBC) localization, classification, and morphological assessment. Despite deep learning advances in medical imaging, leukemia analysis lacks a large, diverse multi-task dataset, while existing small datasets lack domain diversity, limiting real-world applicability. To overcome dataset
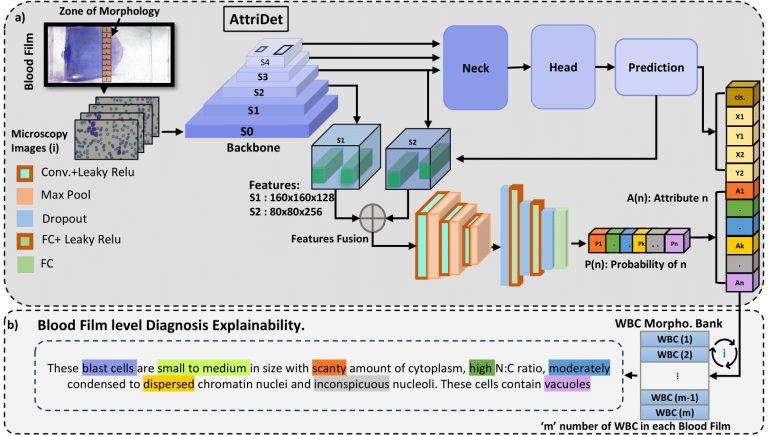
challenges, we present a large-scale WBC dataset named ‘Large Leukemia Dataset’ (LLD) and novel methods for detecting WBC with their attributes. Our contribution here is threefold. First, we present a large-scale Leukemia dataset collected through Peripheral Blood Films (PBF) from several patients, through multiple microscopes, multi-cameras, and multi-magnification. To enhance diagnosis explainability and medical expert acceptance, each leukemia cell is annotated at 100x with 7 morphological attributes, ranging from Cell Size to Nuclear Shape. Secondly, we propose a multi-task model that not only detects WBCs but also predicts their attributes, providing an interpretable and clinically meaningful solution. Third, we propose a method for WBC detection with attribute analysis using sparse annotations. This approach reduces the annotation burden on hematologists, requiring them to mark only a small area within the field of view. Our method enables the model to leverage the entire field of view rather than just the annotated regions, enhancing learning efficiency and diagnostic accuracy. From diagnosis explainability to overcoming domain-shift challenges, presented datasets could be used for many challenging aspects of microscopic image analysis.
Contributions
- Large Leukemia Dataset (LLD) consist of LeukemiaAttri and Sparse-LeukemiaAttri. LeukemiaAttri: A large-scale, multi-domain dataset of 2.4K microscopic images captured using microscopes of varying costs, two cameras, and three resolutions (10x, 40x, 100x). Each image includes WBC localization and morphological attribute annotations for explainable diagnosis. Sparse-LeukemiaAttri: A 40x FoV dataset with sparsely annotated training data (1,546 images) and fully annotated test data (185 images).
- Two detection models are proposed:
– AttriDet is a fully supervised detector designed to identify WBCs and their morphological attributes.
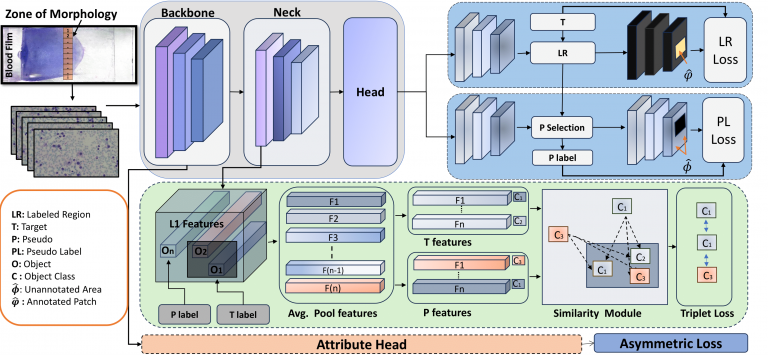
– SLA-Det is a sparse WBC detector that enhances pseudo-label selection by incorporating problem-specific constraints, including size, objectness score, and class label entropy. - Key results 1: SLA-Det processes slides 4x faster than AttriDet at 40x resolution (vs. 100x) while maintaining accuracy. SLA-Det outper forms state-of-the-art sparse detection methods by 34.7 mAP @50, demonstrating superior efficiency and performance.
The work emphasizes domain diversity, diagnostic interpretability, and efficient sparse annotation strategies for real-world leukemia analysis.
Comparison of the proposed dataset with existing leukemia datasets
| Dataset | Type | Multi. Micro. | Multi. Cells in Image | BBX | Multi. Res. | No. of WBC's | WBC Classes | Morphology |
|---|---|---|---|---|---|---|---|---|
| IDB | ALL | ❌ | ✔ | ✔ | ❌ | 510 (LB) | 2 | ❌ |
| IDB2 | ALL | ❌ | ❌ | ❌ | ❌ | 260 | 2 | ❌ |
| LISC | Normal | ❌ | ❌ | ❌ | ❌ | 250 | 5 | ❌ |
| Munich | AML | ❌ | ❌ | ❌ | ❌ | 18,365 | 15 | ❌ |
| Raabin | Normal | ❌ | ✔ | ✔ | ❌ | 17,965 | 5 | ❌ |
| HRLS | Multi. | ❌ | ✔ | ❌ | ❌ | 16,027 | 9 | ❌ |
| WBCAtt | Normal | ❌ | ❌ | ❌ | ❌ | 10,298 | 5 | ✔ |
| LeaukemiaAttri | Multi. | ✔ | ✔ | ✔ | ✔ | 88,294 | 14 | ✔ |
AttriDet
SLA-DET
Sparse-LeukemiaAttri Dataset
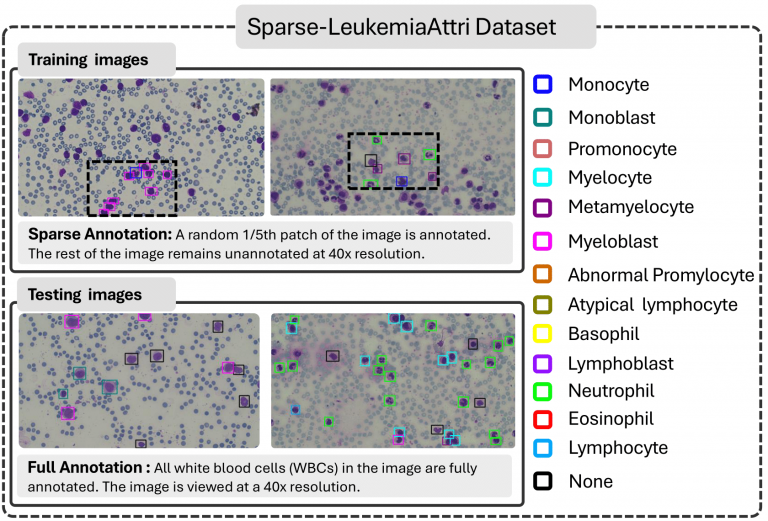
Our Sparse-LeukemiaAttri dataset is a specialized case of sparsely supervised and semi-supervised object detection datasets. Unlike traditional sparsely supervised datasets where annotations are randomly missed, each of our images contains a fully labeled patch. Similarly, while semi-supervised approaches
typically involve fully labeled and fully unlabeled sets, our dataset consists of partially annotated patches within each image, rather than entirely labeled or unlabeled images.
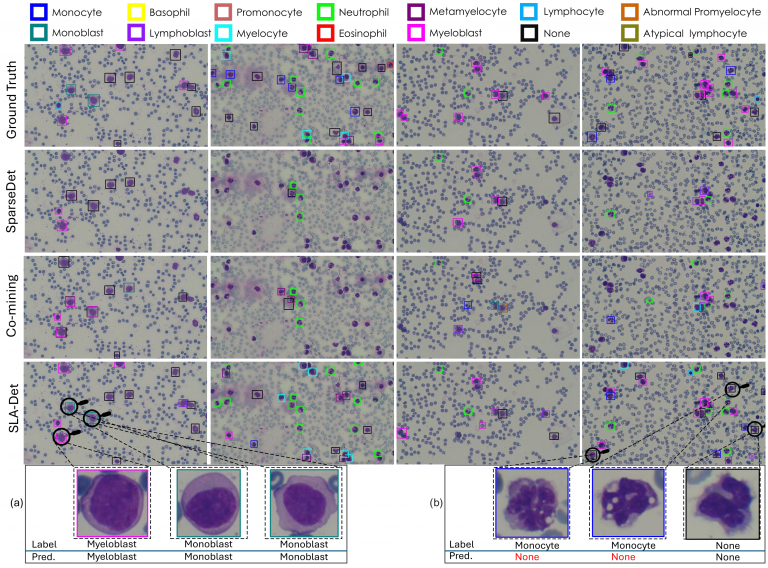
Qualitative Results
The Figure shows that the SLA-Det localization and classification capability significantly outperforms the competitive approaches. Figure (a) and Figure (b) show the zoomed-in version of some of the cells. In Figure (a) all three cells belong to the same myeloid lineage and exhibit nearly identical morphological features. The primary difference between myeloblasts and monoblasts is the nucleus’s position within the cytoplasm: monoblasts
have a nucleus surrounded by a more abundant and vacuolated cytoplasm, while myeloblasts have a more distinct nuclear-cytoplasmic boundary. Despite these minor differences, the detector effectively identifies both cell types. Figure (b) demonstrates a failure case of our approach, where the shapes
of the cell nucleus and cytoplasm are highly irregular. The first two cells are labeled as ‘Monocyte’ and the third cell is labeled as ‘None’. We believe that the minor staining effects might be the reason to confuse the detector, potentially resulting in incorrect classifications.
About Authors

Abdul Rehman

Talha
Meraj

Aiman Mahmood

Ayesha
Imran

Mohsen Ali

Waqas Sultani

Mubarak Shah
@article{rehman2024leveraging,
title={Leveraging Sparse Annotations for Leukemia Diagnosis
on the Large Leukemia Dataset},
author={Rehman, Abdul and Meraj, Talha and Minhas, Aiman Mahmood and Imran, Ayisha and Ali, Mohsen and Sultani, Waqas},
journal={bioRxiv},
pages={2024--11},
year={2024},
publisher={ABC}
} 


