Abstract
Malaria, a fatal but curable disease claims hundreds of thousands of lives every year. Early and correct diagnosis is vital to avoid health complexities, however, it depends upon the availability of costly microscopes and trained experts to analyze blood-smear slides. Deep learning-based methods have the potential to not only decrease the burden of experts but also improve diagnostic accuracy on low-cost microscopes. However, this is hampered by the absence of a reasonable size dataset. One of the most challenging aspects is the reluctance of the experts to annotate the dataset at low magnification on low-cost microscopes. We present a dataset to further the research on malaria microscopy over the low-cost microscopes at low magnification. Our large-scale dataset consists of images of blood-smear slides from several malaria-infected patients, collected through microscopes at two different cost spectrums and multiple magnifications. Malarial cells are annotated for the localization and life-stage classification task on the images collected through the high-cost microscope at high magnification. We design a mechanism to transfer these annotations from the high-cost microscope at high magnification to the low-cost microscope, at multiple magnifications. Multiple object detectors and domain adaptation methods are presented as the baselines. Furthermore, a partially supervised domain adaptation method is introduced to adapt the object-detector to work on the images collected from the low-cost microscope.
Our Contribution
- We collect the first large-scale multi-microscope multi-magnification malarial image dataset from the thin-blood smear slides. Dataset has been annotated by the medical expert with more than 20 years of experience, for both the malarial cell localization and life-stage classification tasks.
- An annotation mechanism (consisting of multiple mobile cameras and microscopes) is designed that records the microscope’s x-y transnational control knobs positions to speed up the data collection, handles issues of calibrations across magnifications and also across microscopes.
- We compute the baseline results of several object detectors and domain adaptation methods on our dataset, creating a benchmark for the computer-aided malaria detection task.
- Finally, utilizing the problem-specific constraints, a partially supervised domain adaptation mechanism is designed.
Dataset Details
| Microscope Type | No. of Images | No. of Annotated Malarial Cells |
|---|---|---|
| HCM (High Cost Microscope) | 1257 x 3 = 3771 | 3,624 x 3 = 10,872 |
| LCM (Low Cost Microscope) | 1257 x 3 = 3771 | 3,153 x 3 = 9459 |
Our dataset consists of 1257 images on three different magnifications (100x, 400x, and 1000x) and on two different microscopes of different qualities and costs. Our dataset is first of its kind, consisting images from same locations of thin blood smears on three different magnification scales and on two different microscopes. It was a real challenge to trace and capture images of same locations of thin blood smears on two different microscopes and across different magnifications. We have fewer number of annotated malarial cells on LCM as compared to HCM as the field of view of LCM is smaller than the field of view of HCM.
Comparison with Existing Datasets
| Malaria Dataset | Across Microscopes | Multi Magnification | No. of Malarial Cells | BBox Localization | No. of Images |
|---|---|---|---|---|---|
| BBBC041 | N0 | No | 2452 | Yes | 1364 |
| Malaria655 | No | No | 557 | No | 655 |
| MPIDB | No | No | 840 | No | 229 |
| IML | No | No | 529 | Yes | 345 |
| Ours | Yes | Yes | 20,331* | Yes | 7542* |
Medical expert traversed multiple slides identifying 1257 malarial regions using HCM at 1000x. Images corresponding to these locations were collected at all the magnifications in both HCM and LCM microscopes resulting in the dataset of size 7,542 (=2 x 3 x 1257) and
total number of malarial cells 3 x 3,624 (HCM) + 3 x 3,153 (LCM) = 20,331.
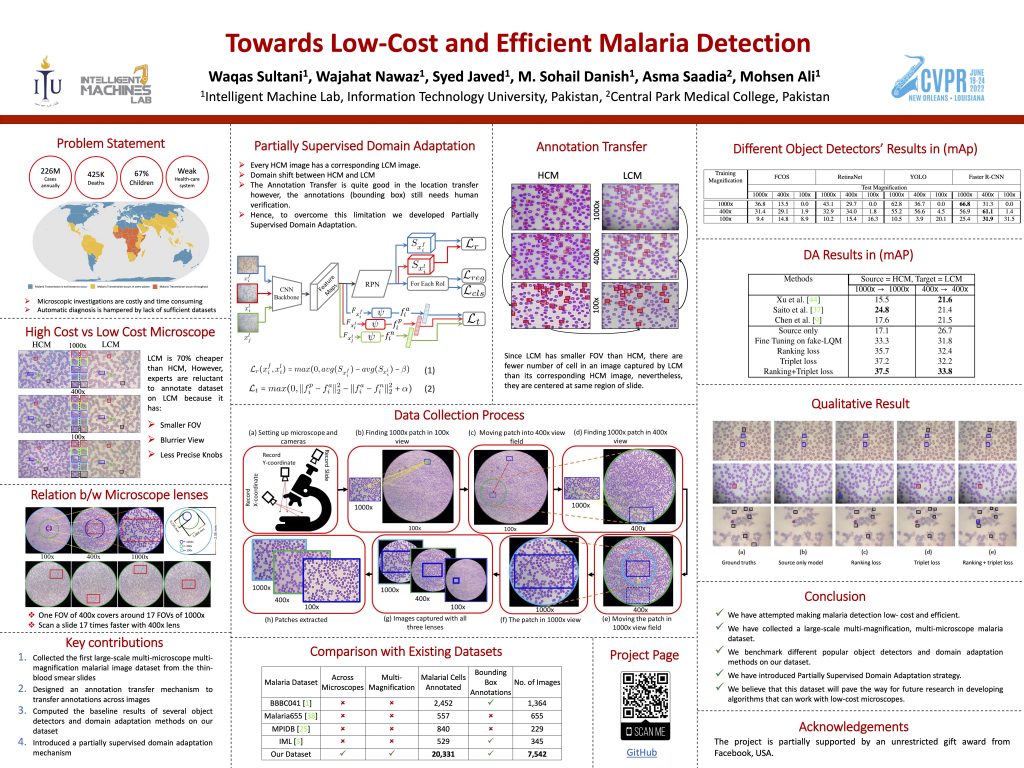
Desktop Application for Dataset Collection
We developed a desktop application that helps trace and capture images of particular regions of thin blood smear slides with different microscopes on different magnifications.
Dataset Annotation and Annotation Transfer Mechanism

The dataset was annotated on 1000x images and these annotations were transfered to images of different magnifications and different microscopes using homography and annotations for each image were verified and rectified one by one manually.
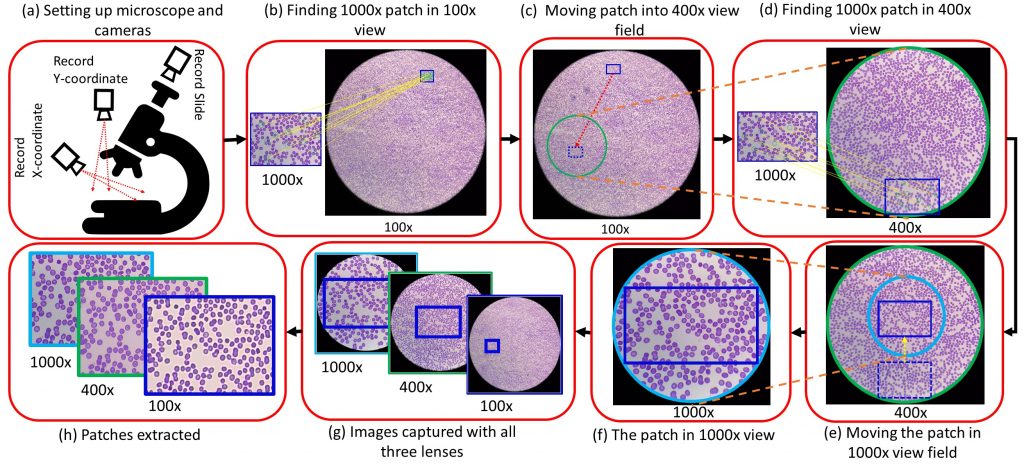
Our Partially Supervised Domain Adaptation Mechanism
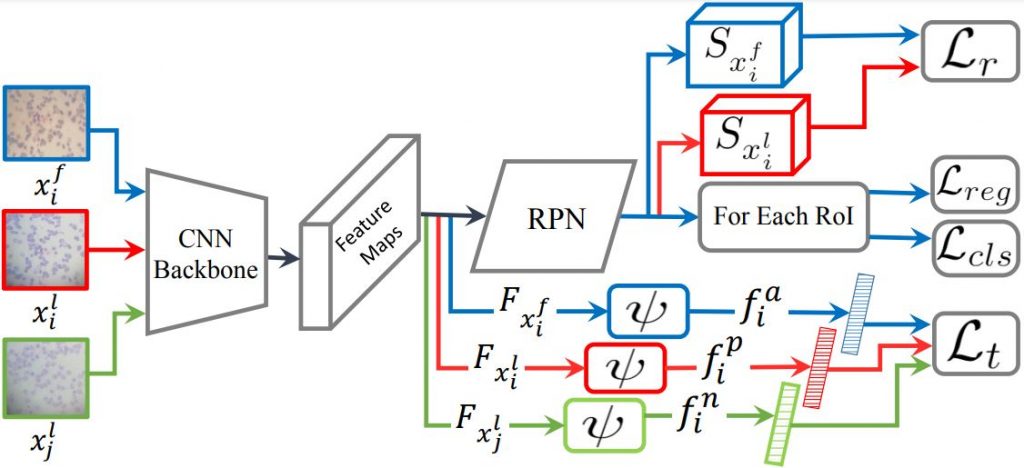
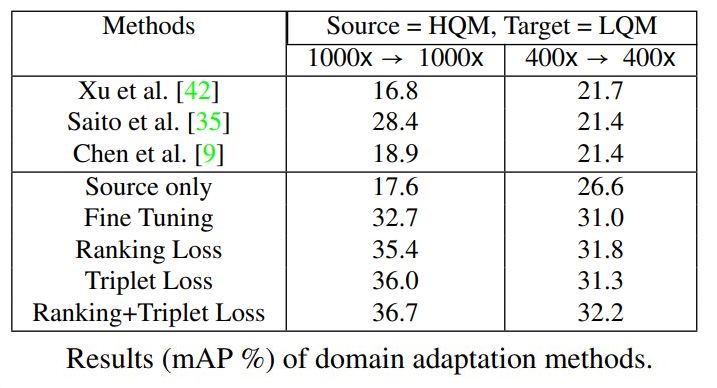
Baseline Results of Several Object Detectors and Domain Adaptation Methods on Our Dataset
Qualitative Results
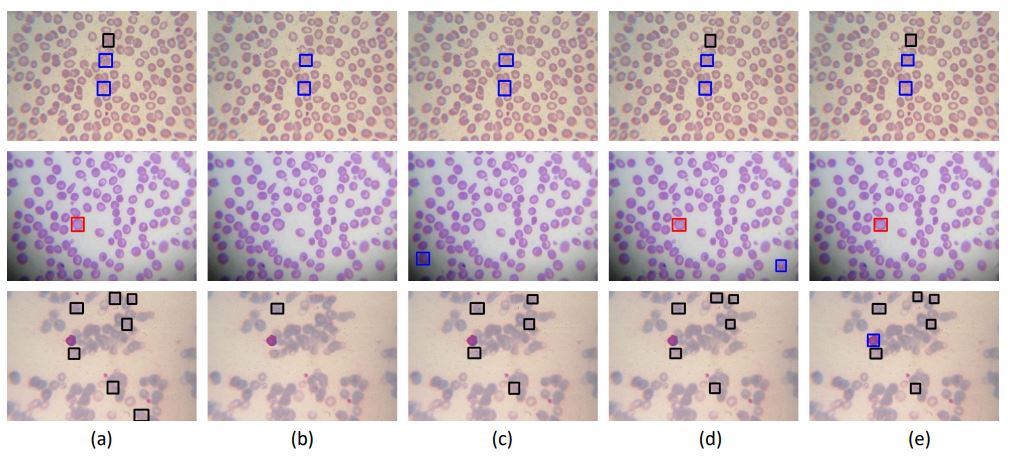
Red, black and blue squares show gametocyte, ring, and trophozoite respectively. (a) shows ground truths, (b)
shows the results of source only model on LCM, (c) shows results when model trained on HCM, fine-tuned on fake LCM with ranking
loss. (d) shows results when model trained on HCM, fine-tuned on fake LCM with triplet loss, and (e) shows the results of (c) + (d).
Citation
If you are using our dataset, please cite this publication.
@InProceedings{Sultani_2022_CVPR, author = {Sultani, Waqas and Nawaz, Wajahat and Javed, Syed and Danish, Muhammad Sohail and Saadia, Asma and Ali, Mohsen},
title = {Towards Low-Cost and Efficient Malaria Detection},
booktitle = {Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)},
month = {June},
year = {2022},
pages = {20687-20696} }